Jarrett
Educational Background
Joe Jarrett received a B.S. in chemistry from the University of Michigan in 1988. He obtained a Ph.D. in Biochemistry from the Department of Chemistry at the Massachusetts Institute of Technology in 1993, working with Peter T. Lansbury, Jr. on the mechanism of amyloid fibril formation in Alzheimer’s disease. He then returned to the University of Michigan for postdoctoral studies in the laboratory of Rowena G. Matthews, focusing on mechanistic studies of cobalamin-dependent methionine synthase. He was a member of the faculty of the Department of Biochemistry and Biophysics at the University of Pennsylvania from 1997 to 2005. He joined the Department of Chemistry at the University of Hawaii in 2006.

- Bockman, M. R.; Engelhart, C. A.; Cramer, J. D.; Howe, M. D.; Mishra, N. K.; Zimmerman, M.; Larson, P.; Alvarez-Cabrera, N.; Park, S. W.; Boshoff, H. I. M.; Bean, J. M.; Young, V. G., Jr.; Ferguson, D. M.; Dartois, V.; Jarrett, J. T.; Schnappinger, D.; Aldrich, C. C., Investigation of ( S)-(-)-Acidomycin: A Selective Antimycobacterial Natural Product That Inhibits Biotin Synthase. ACS infectious diseases 2019, 5, 598-617.
- Jarrett, J. T., Surprise! A Hidden B12 Cofactor Catalyzes a Radical Methylation. J. Biol. Chem. 2019, 294, 11726-11727.
- Tao, L.; Stich, T. A.; Fugate, C. J.; Jarrett, J. T.; Britt, R. D., EPR-Derived Structure of a Paramagnetic Intermediate Generated by Biotin Synthase BioB. J. Am. Chem. Soc. 2018, 140, 12947-12963.
- Cramer, J. D.; Jarrett, J. T., Purification, Characterization, and Biochemical Assays of Biotin Synthase From Escherichia coli. Methods Enzymol. 2018, 606, 363-388.
- Jarrett, J. T., The Biosynthesis of Thiol- and Thioether-containing Cofactors and Secondary Metabolites Catalyzed by Radical S-Adenosylmethionine Enzymes. J. Biol. Chem. 2015, 290, 3972-3979.
Research Interests
The S-Adenosylmethionine-Dependent Radical Enzymes
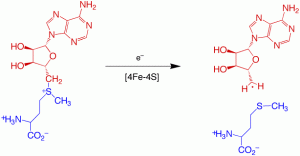
Organic radicals are used by a number of enzymes to catalyze biochemical transformations with high-energy barriers that would be difficult to accomplish through non-radical heterolytic chemistry. Well known examples include the reduction of an alcohol to an alkane catalyzed by ribonucleotide reductase or carbon chain rearrangements catalyzed by methylmalonyl CoA mutase or glutamate mutase. Organic radicals can be generated in enzymes through only three general schemes: metal-activated oxygen chemistry, adenosylcobalamin chemistry, or reduction of the sulfonium of S-adenosylmethionine (AdoMet).
Enzymes that catalyze the reduction of AdoMet all contain a [4Fe-4S] cluster that interacts covalently with the methionyl portion of AdoMet. This interaction likely plays an important role in promoting electron transfer into the relatively low-potential sulfonium. We are interested in understanding how the electronic structure of the FeS cluster and the sulfonium are perturbed by this interaction, and whether electron transfer and sulfonium reduction requires a structurally specific interaction or simply close proximity.
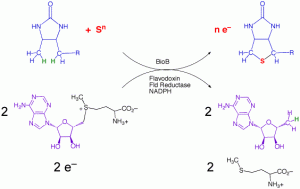
The AdoMet Radical Enzyme Biotin Synthase
Biotin (vitamin H) is synthesized in microbes and plants through a conserved biosynthetic pathway that generates dethiobiotin from L-alanine, pimeloyl CoA, CO2, and NH3 (from AdoMet). The final step in the pathway requires the substitution of sulfur for hydrogen at the C6 and C9 positions to create the thiophane ring. The enzyme that catalyzes this step in the reaction contains iron-sulfur clusters and is an AdoMet-dependent radical enzyme. The overall reaction catalyzed is:
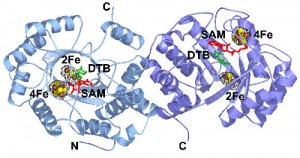
In collaboration with Dr. Cathy Drennan (MIT), we have determined the structure of the enzyme from E. coli with substrates bound. On the basis of this structure, and related mechanistic studies, we have proposed that the biotin thiophane sulfur is derived from a [2Fe-2S] cluster bound within the core of the enzyme. Current studies are focused on obtaining spectroscopic evidence for a dethiobiotin-FeS cluster covalent intermediate and understanding the role of conserved protein residues in guiding this transformation.
In Vivo Iron-Sulfur Cluster Assembly
Although simple iron-sulfur clusters are spontaneously assembled in solution, virtually all known organisms contain specific proteins that function to guide the controlled assembly of [2Fe-2S] and [4Fe 4S] clusters. One of the most conserved systems for iron-sulfur cluster assembly, the ISC system, is contained within a single genetic operon in E. coli that codes for 7 proteins: a pyridoxal phosphate-dependent cysteine desulfurase that generates S0 as persulfide sulfur destined for cluster assembly (IscS), two possible scaffold proteins that can bind either [2Fe-2S], [4Fe 4S], or Fe2+/3+ under various experimental conditions (IscU and IscA), a ferredoxin for redox control (Fdx), and an ATP-dependent chaperone/co-chaperone pair that may assist in folding/unfolding or cluster transfer (HscA and HscB).
We have found that the chaperone HscA and the scaffold IscU will form a complex with several apoproteins that are targets for FeS cluster transfer. Our current studies are focused on cluster assembly in biotin synthase, which is stably folded in the apoprotein state, and where correct cluster assembly can be distinguished from indiscriminant assembly by the ability of the resulting protein to form biotin. Current studies focus on using spectroscopic methods to characterize the state of the FeS cluster prior to and during the cluster transfer process.
Potential projects:
- Model studies of sulfonium reduction chemistry.
- Re-engineering biotin synthase to contain a spectroscopic probe to follow AdoMet cleavage and radical generation.
- Mutagenesis of conserved protein residues in biotin synthase to probe the role of the enzyme in guiding the radical chemistry.
- Use of deuterated dethiobiotin to determine isotope effects for the cleavage of C-H bonds by biotin synthase, to better understand the transition state structure.
- Spectroscopic and electrochemical studies of cluster assembly and transfer within protein members of the ISC system.
- Mutagenesis of conserved protein residues in ISC proteins to probe the role of the proteins in controlling cluster assembly and targeting to specific apoproteins.